Genome Instability. Population Dynamics. Drug Resistance. Evolution.
University of Minnesota Medical School
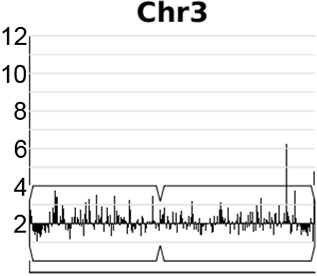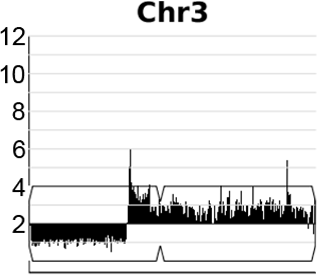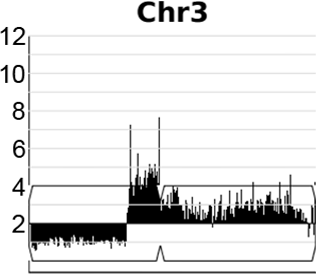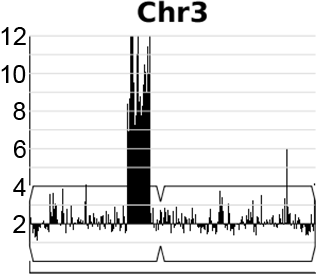
A dynamic, expandable and reversible, segmental copy number amplification on chromosome 3 in Candida albicans promotes acquisition of drug resistance.
Video generated from data in Todd & Selmecki, eLife, 2020
Research
Understanding the dynamics of how growth-promoting mutations arise and accumulate in a population of cells is a fundamental problem underlying our understanding of drug resistance, tumorigenesis, and the treatment of cancer. We use experimental evolution, mathematical modeling, and comparative genomics to understand the impact of mutations on the adaptation of a cell and its surrounding population.
We employ diverse yeast model systems (Saccharomyces cerevisiae, Candida albicans, Candida auris, etc.) to understand how genome instability contributes to adaptation (eg. antifungal drug resistance) and determine the underlying mechanisms that promote genome instability.
In the human fungal pathogen Candida albicans, genome rearrangements resulting in copy number variation (CNV) and loss of heterozygosity (LOH) confer increased virulence and antifungal drug resistance, yet the mechanisms driving these rearrangements are not completely understood. We recently identified an extensive array of long repeat sequences (65–6499 bp) that are associated with CNV, LOH, and chromosomal inversions and are a significant source of genome plasticity across diverse strain backgrounds - including clinical, environmental, and experimentally evolved C. albicans isolates. Many of these long repeat sequences were uncharacterized and encompass one or more coding sequences that are actively transcribed. Further, the repeats associated with genome rearrangements were predominantly inverted and separated by up to ~1.6 Mb, an extraordinary distance for homology-based DNA repair/recombination in yeast!
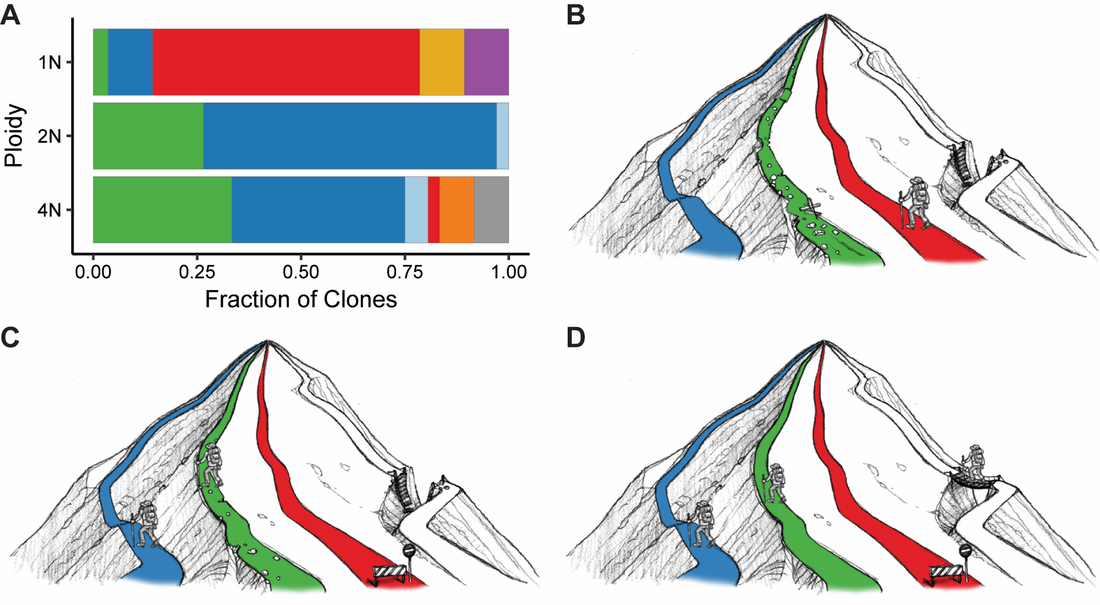
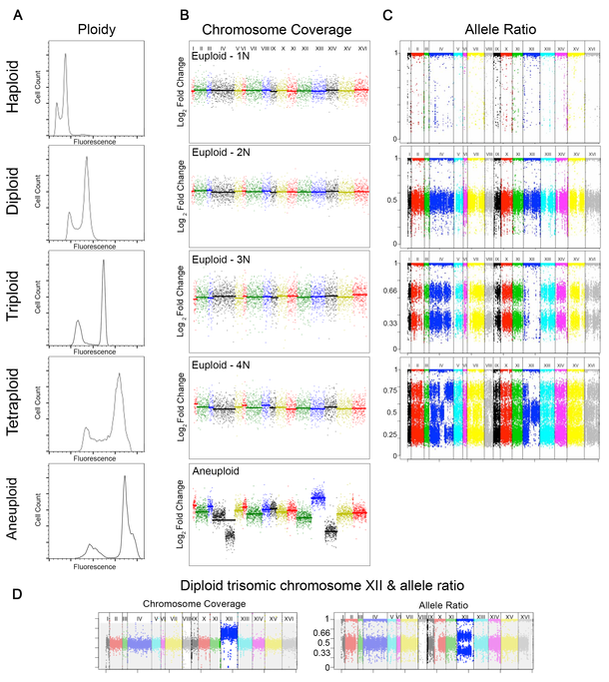
We also utilize flow cytometry-based systems that enable us to detect the acquisition and spread of beneficial mutations within populations. We found that polyploid S. cerevisiae adapted more rapidly than isogenic haploid or diploid cells in poor carbon medium, and that polyploid cells acquired more mutations, including point mutations, large segmental aneuploidies, and whole chromosome aneuploidies (Selmecki et al., Nature 2015). Additionally, polyploid cells acquired a broader spectrum of beneficial mutations than lower ploidy cells (Scott et al., MBE 2017). We continue to use these ploidy lineages to study how changes in chromosome number (ploidy and aneuploidy), cell size, and environment affect genome stability and evolvability.
Our previous research identified chromosome aneuploidy as a driver for the acquisition of antifungal drug resistance in the pathogenic yeast C. albicans (Selmecki et al., Science 2006). We found that aneuploid cells arose within a population very rapidly in the presence of antifungal drug (Selmecki et al., PLoS Genetics 2009), and that increased copy number of two specific genes found on the most common aneuploid chromosome provided the drug resistance phenotype (Selmecki et al., Molecular Microbiology 2008).
We employ diverse yeast model systems (Saccharomyces cerevisiae, Candida albicans, Candida auris, etc.) to understand how genome instability contributes to adaptation (eg. antifungal drug resistance) and determine the underlying mechanisms that promote genome instability.
In the human fungal pathogen Candida albicans, genome rearrangements resulting in copy number variation (CNV) and loss of heterozygosity (LOH) confer increased virulence and antifungal drug resistance, yet the mechanisms driving these rearrangements are not completely understood. We recently identified an extensive array of long repeat sequences (65–6499 bp) that are associated with CNV, LOH, and chromosomal inversions and are a significant source of genome plasticity across diverse strain backgrounds - including clinical, environmental, and experimentally evolved C. albicans isolates. Many of these long repeat sequences were uncharacterized and encompass one or more coding sequences that are actively transcribed. Further, the repeats associated with genome rearrangements were predominantly inverted and separated by up to ~1.6 Mb, an extraordinary distance for homology-based DNA repair/recombination in yeast!
We also utilize flow cytometry-based systems that enable us to detect the acquisition and spread of beneficial mutations within populations. We found that polyploid S. cerevisiae adapted more rapidly than isogenic haploid or diploid cells in poor carbon medium, and that polyploid cells acquired more mutations, including point mutations, large segmental aneuploidies, and whole chromosome aneuploidies (Selmecki et al., Nature 2015). Additionally, polyploid cells acquired a broader spectrum of beneficial mutations than lower ploidy cells (Scott et al., MBE 2017). We continue to use these ploidy lineages to study how changes in chromosome number (ploidy and aneuploidy), cell size, and environment affect genome stability and evolvability.
Our previous research identified chromosome aneuploidy as a driver for the acquisition of antifungal drug resistance in the pathogenic yeast C. albicans (Selmecki et al., Science 2006). We found that aneuploid cells arose within a population very rapidly in the presence of antifungal drug (Selmecki et al., PLoS Genetics 2009), and that increased copy number of two specific genes found on the most common aneuploid chromosome provided the drug resistance phenotype (Selmecki et al., Molecular Microbiology 2008).
Recently Published
Expandable and reversible copy number amplification drives rapid adaptation to antifungal drugs eLife (2020) Robert Todd and Anna Selmecki
Genome Plasticity in Candida albicans is Driven by Long Repeat Sequences
eLife (2019) Robert Todd, Tyler Wikoff, Anja Forche, Anna Selmecki
eLife (2019) Robert Todd, Tyler Wikoff, Anja Forche, Anna Selmecki
The Influence of Polyploidy on the Evolution of Yeast Grown in a Sub-optimal Carbon Source.
Molecular Biology and Evolution (2017) Amber Scott, Phillip Richmond, Robin Dowell, Anna Selmecki
#OpenAccess
Ploidy Variation in Fungi: Polyploidy, Aneuploidy, and Genome Evolution.
Microbiology Spectrum. (2017) Robert T. Todd, Anja Forche, Anna Selmecki
Microbiology Spectrum. (2017) Robert T. Todd, Anja Forche, Anna Selmecki
Our chapter in the book Fungal Kingdom
Chapter 28: Ploidy Variation in Fungi: Polyploidy, Aneuploidy, and Genome Evolution.
(2017) Robert T. Todd, Anja Forche, Anna Selmecki
Chapter 28: Ploidy Variation in Fungi: Polyploidy, Aneuploidy, and Genome Evolution.
(2017) Robert T. Todd, Anja Forche, Anna Selmecki